Projects
Here are some of the things I’ve been working on.
MolE enables antimicrobial discovery.

In this project, we developed a self-supervised deep learning model that learns a representation of molecular structures that captures important chemical and topological features. We apply this framework to the task of antimicrobial discovery, where we show that our model can be used to predict antimicrobial activity of molecules with high accuracy. Using our model’s predictions, we identified new antimicrobial compounds that were experimentally confirmed to be active against the human pathogen Staphylococcus aureus.
Read the paper published in Nature Communications
GitHub repositories:
- MolE.
- mole_antimicrobial_potential.
DGrowthR: Comprehensive analysis of bacterial growth.
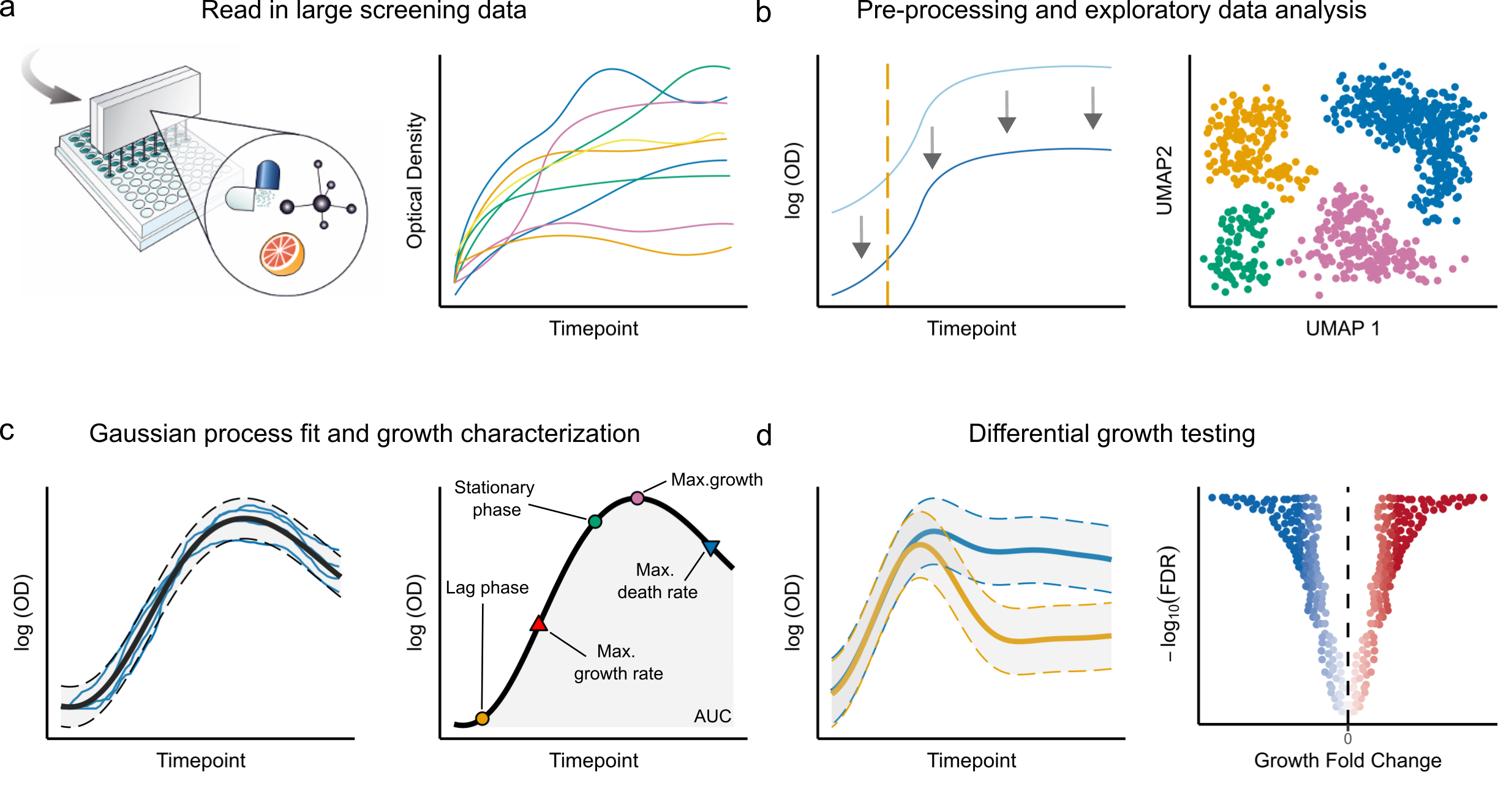
In this project, we developed a comprehensive framework for the analysis of bacterial growth data, based on Gaussian Process regression. Without the need for any prior assumptions about the functional form of the growth curve, our method can accurately estimate growth parameters, provides options for exploratory data analysis, and a statistical testing framework to compare growth curves across different conditions. Using this tool we analyzed multiple datasets, uncovering and extending previous findings on the growth of bacteria in response to chemical perturbations.
Read the pre-print.
GitHub links:
Statistical modeling of bacterial stress response.
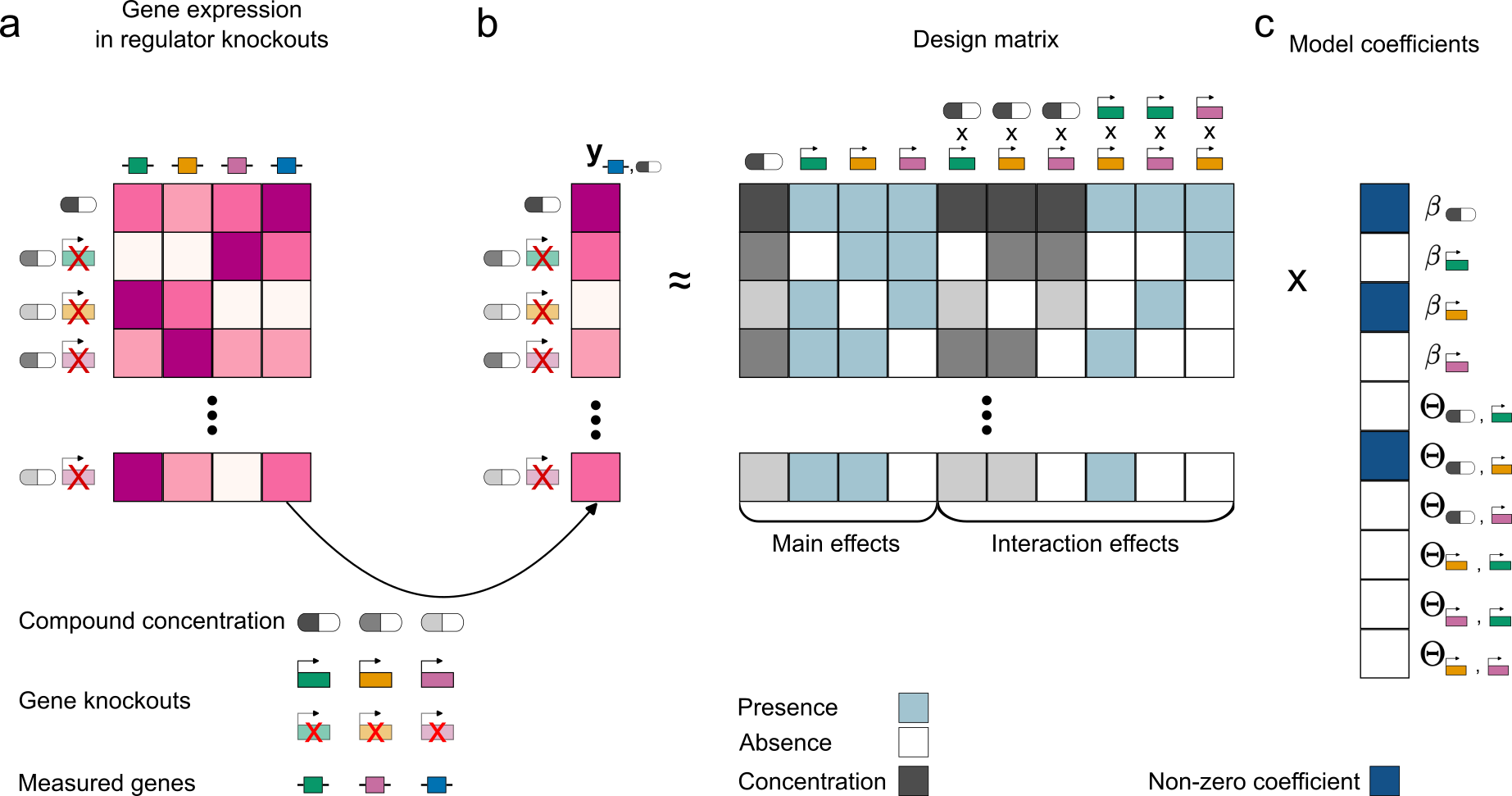
Here, we created a statistical model that analyzes data from chemical-genetic screens to identify the regulatory relationships that control bacterial stress response. We developed a hierarchical interaction model that captures regulator contributions to the observed changes in gene expression in Escherichia coli in response to chemical perturbations. Using this model, we identified the regulatory networks responsible for antibiotic antagonism mediated by Caffeine. This project was done in close collaboration with Christoph Binsfeld and Dr. Ana Rita Brochado.
Read the paper now accepted in Plos Biology.
Bacterial Single-Cell RNA-Seq analysis.
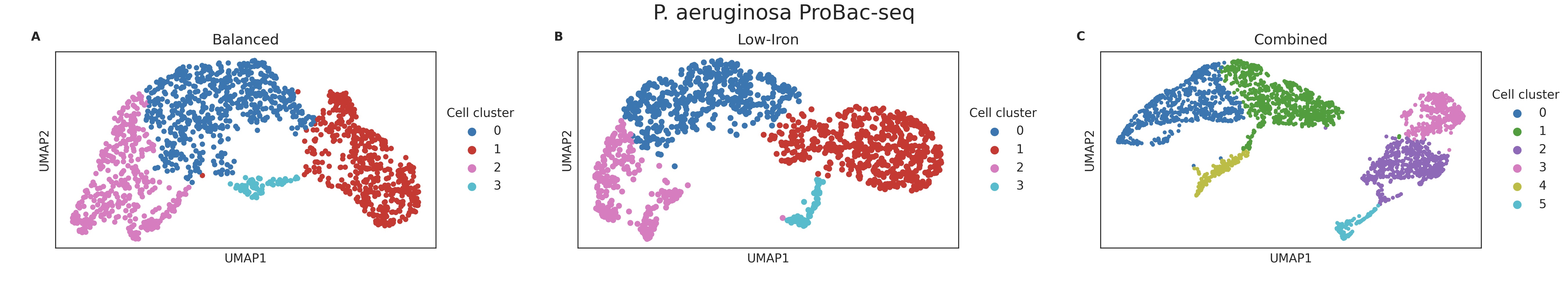
In this project, led by Johannes Ostner, we developed a general workflow for the analysis of bacterial single-cell RNA sequencing data. We created a set of tools that allow for quality control, normalization, clustering, and differential expression analysis. We applied this workflow to analyze single-cell RNA-seq data from Bacillus subtilis, Pseudomonas aeruginosa, and Klebsiella pneumoniae.
Read the pre-print.
StressRegNet
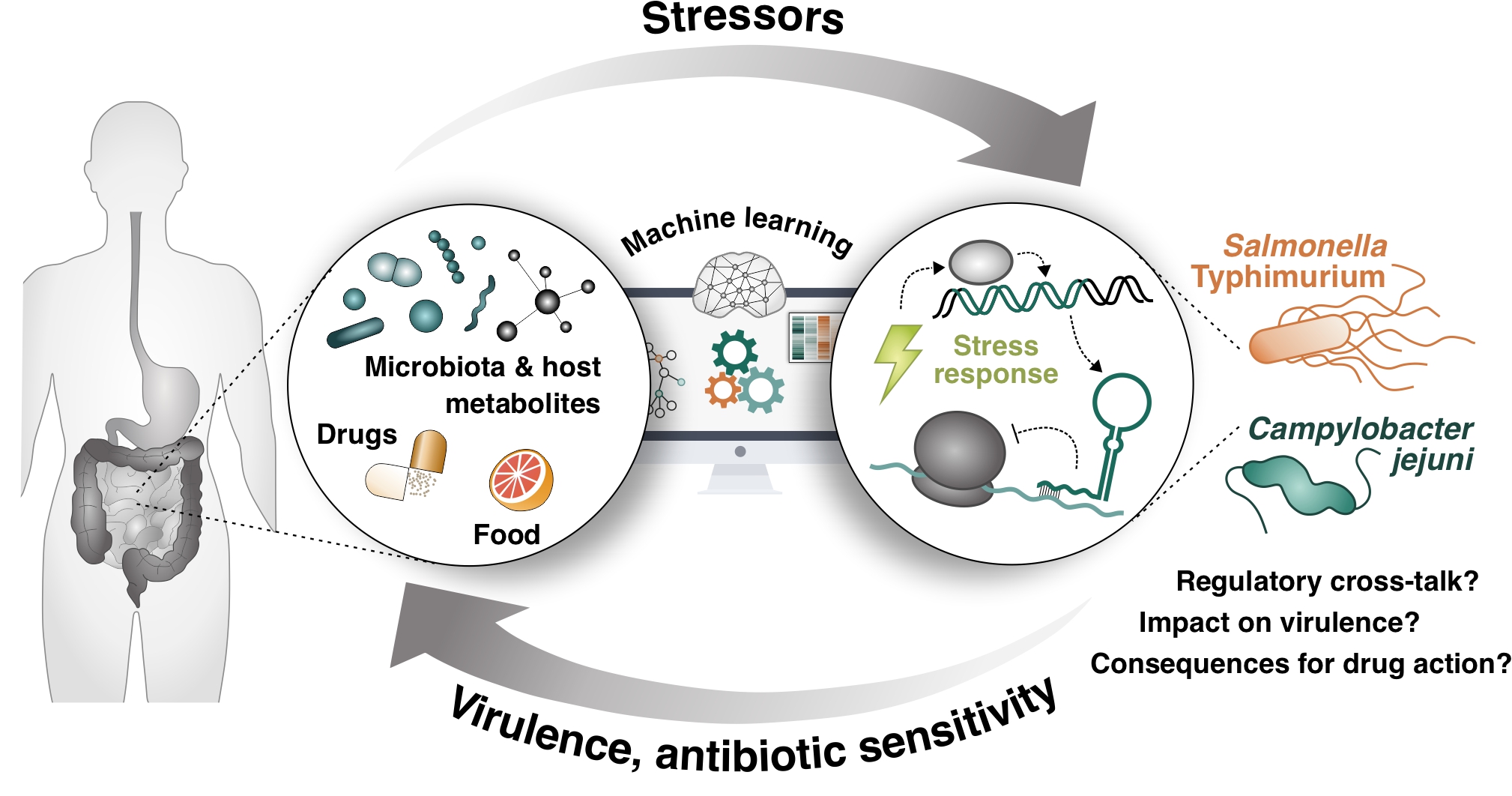
Here, we aim to identify chemical stressors and the bacterial regulatory pathways that control host adaptation and antibiotic sensitivity. By using a high-throughput chemical genomics approach, we will explore chemical stimuli from the host environment that induce stress response pathways in Salmonella and Campylobacter, and elucidate the underlying molecular crosstalk between sensory pathways of these microbes.
This project is part of the bavarian Bayresq research consortium, and is done in close colaboration with members of Dr. Cynthia Sharma and Dr. Ana Rita Brochado research groups.
Non-Academic Projects
I also like to dabble on other types of analysis. Here are some things I’ve done for fun.
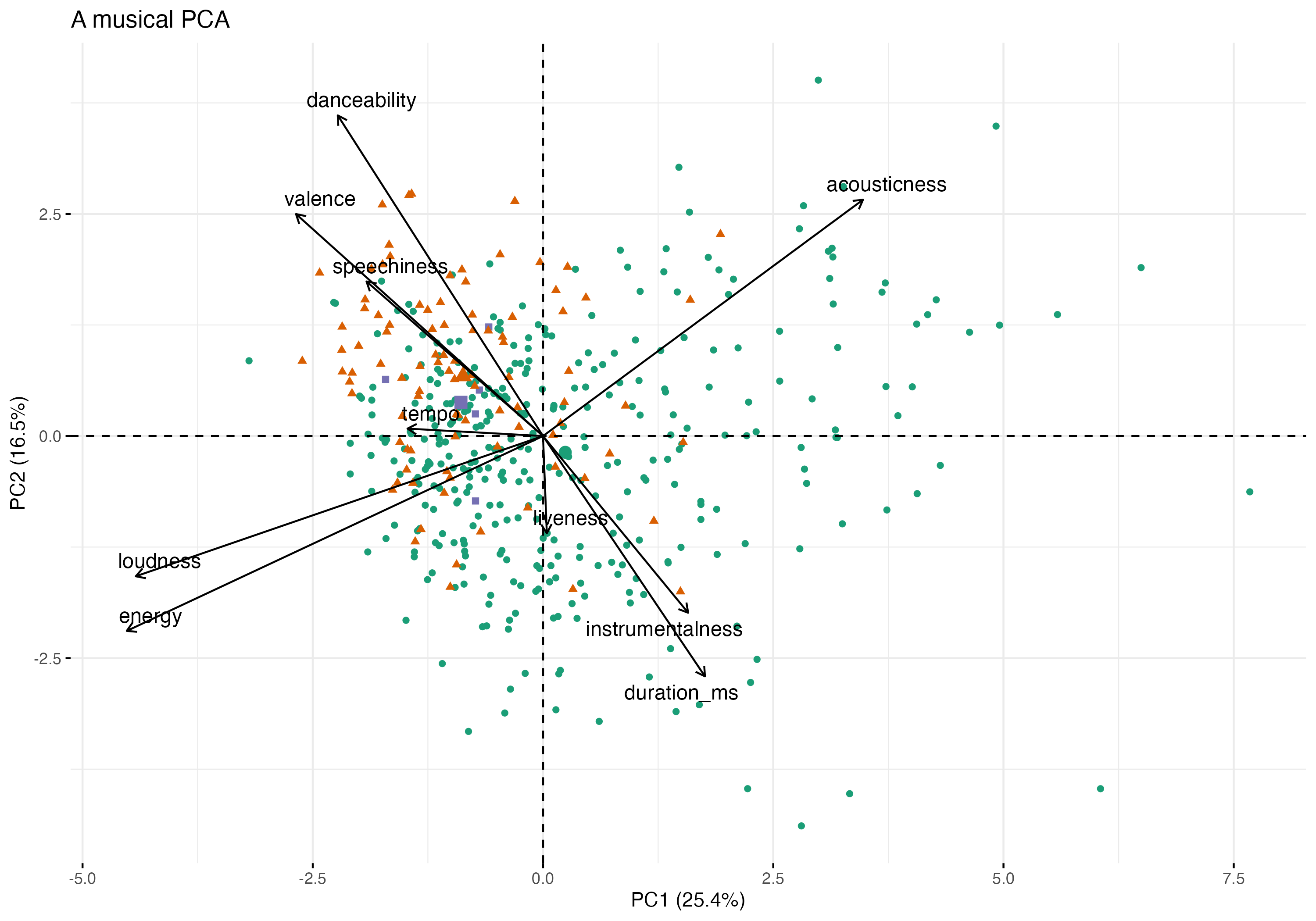
Comparing Musical taste with Spotify.